| structure name | ASYMMETRIC DNA-BENDING IN THE CRE-LOXP SITE-SPECIFIC RECOMBINATION SYNAPSE (PROTEIN BACTERIOPHAGE P1 CRE GENE) |
| reference | Van Duyne et al. |
| source | Enterobacteria phage P1 |
| experiment | X-ray (resolution=2.70, R-factor=0.227) |
| structural superfamily | lambda integrase-like, N-terminal domain;DNA breaking-rejoining enzymes; |
| sequence family | Phage integrase family; |
| multimeric complexes | 1kbu_AB 1ma7_AB 1nzb_AB 1ouq_AB 1ouq_EF 1pvp_AB 1pvq_AB 1pvr_AB 1q3v_AB 1q3v_EF 1xo0_AB 2hof_AB 2hoi_GH 3c28_AB 3c29_AB 3mgv_AB 3mgv_CD 4crx_AB 5crx_AB 7rhx_AB 7rhz_AB |
| reference complex | 7rhx_G |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | HTMSKTQQKRRKSRER |
Estimated binding specificities ?
| readout + contact | 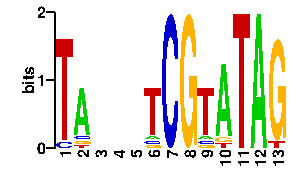 |
A | 0 75 24 24 24 6 0 0 6 82 0 96 0 C | 7 6 24 24 24 6 96 0 6 2 0 0 0 G | 0 9 24 24 24 6 0 96 6 6 0 0 90 T | 89 6 24 24 24 78 0 0 78 6 96 0 6scan! |
home
updated Mon Dec 18 12:25:27 2023
