| structure name | MUTM SET 1 GPG (Formamidopyrimidine-DNA glycosylase) |
| reference | Verdine et al. J.Biol.Chem. 287 18044 2012 |
| source | Geobacillus stearothermophilus |
| experiment | X-ray (resolution=1.60, R-factor=0.193) |
| structural superfamily | N-terminal domain of MutM-like DNA repair proteins;S13-like H2TH domain;Glucocorticoid receptor-like DNA-binding domain; |
| sequence family | FormamidopyrimidineDNA glycosylase N; FormamidopyrimidineDNA glycosylase H2; Zinc finger found in FPG and IleRS; |
| redundant complexes | 1l1t_A
1l1z_A
1l2b_A
1l2c_A
1l2d_A
1r2y_A
1r2z_A
2f5n_A
2f5o_A
2f5p_A
2f5q_A
2f5s_A
3gp1_A
3gpp_A
3gpy_A
3gq4_A
3gq5_A
3jr4_A
 3jr5_A
3sar_A
3sas_A
3sat_A
3sau_A
3sav_A
3saw_A
3sbj_A
3u6c_A
3u6d_A
3u6e_A
3u6l_A
3u6m_A
3u6o_A
3u6q_A
3u6s_A
4g4n_A
4g4o_A
4g4q_A
4g4r_A 3jr5_A
3sar_A
3sas_A
3sat_A
3sau_A
3sav_A
3saw_A
3sbj_A
3u6c_A
3u6d_A
3u6e_A
3u6l_A
3u6m_A
3u6o_A
3u6q_A
3u6s_A
4g4n_A
4g4o_A
4g4q_A
4g4r_A
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RMRFR |
Estimated binding specificities ?
| contact | 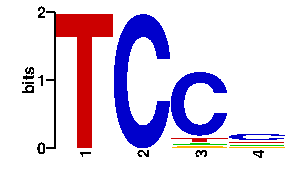 |
A | 0 0 4 15 C | 0 96 81 49 G | 0 0 4 15 T | 96 0 7 17scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: MUTM | ||
| CTGGC | PubMed | 3gpp_A(1.34e-04), 3u6s_A(1.35e-04), 3sau_A(1.34e-04), ... (total=4) |
Dendrogram of similar interfaces ?
matrix format--RGMG----RCFC--RG L +3u6p_A +-1 --RA------RGFT--RA +-2 +3gpu_A ! ! --RTMT----RCFA--RT +-----------3 +6fl1_A ! ! RT--MG----RCFC--RT L --6 +-6tc9_A ! --QALGYA--------RG ! +---------1k3x_A +---5 ----MG--ITRCFGHGRG L ! +------5itu_A +--4 ----MG--ITRCFG--RG +------6lwd_G |
home
updated Tue Dec 19 08:16:49 2023
