| structure name | MUTM LESION RECOGNITION CONTROL COMPLEX WITH N174C CROSSLINKING SITE (DNA glycosylase) |
| reference | Verdine et al. J.Biol.Chem. 285 1468 2010 |
| source | Geobacillus stearothermophilus |
| experiment | X-ray (resolution=1.70, R-factor=0.166) |
| structural superfamily | N-terminal domain of MutM-like DNA repair proteins;S13-like H2TH domain;Glucocorticoid receptor-like DNA-binding domain; |
| sequence family | FormamidopyrimidineDNA glycosylase N; FormamidopyrimidineDNA glycosylase H2; Zinc finger found in FPG and IleRS; |
| reference complex | 3u6p_A |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RMERFRR |
Estimated binding specificities ?
| readout + contact | 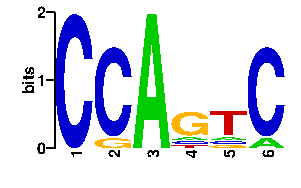 |
A | 0 0 96 10 10 10 C | 96 85 0 10 10 86 G | 0 10 0 66 10 0 T | 0 1 0 10 66 0scan! |
home
updated Mon Dec 18 08:09:44 2023
