| structure name | CRYSTAL STRUCTURE OF A MUTANT IHF (BETAE44A) COMPLEXED WITH THE NATIVE H' SITE (INTEGRATION HOST FACTOR ALPHA-SUBUNIT) |
| reference | Gardner et al. J.Mol.Biol. 330 493 2003 |
| source | ESCHERICHIA COLI |
| experiment | X-ray (resolution=1.95, R-factor=0.232) |
| structural superfamily | IHF-like DNA-binding proteins; |
| sequence family | Bacterial DNAbinding protein; |
| multimeric complexes | 1ihf_AB 1ouz_AB 1owf_AB 1owg_AB 2ht0_AB |
| redundant complexes | 1ihf_A
 1ouz_A
1owg_A
2ht0 1ouz_A
1owg_A
2ht0
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RRNPKI |
Estimated binding specificities ?
| contact | 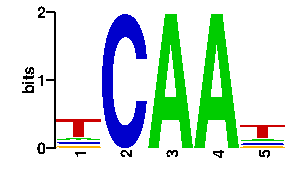 |
A | 56 0 0 0 60 C | 13 0 0 0 12 G | 14 0 0 96 12 T | 13 96 96 0 12scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: IHF | ||
| TTGANCAAATC | PubMed | 1owf_A(5.58e-03) |
| AAAATTGTACA | PubMed | 1owf_A(5.59e-03) |
| TGTACAATTTT | PubMed | 1owf_A(5.59e-03) |
Dendrogram of similar interfaces ?
matrix format------RTRTNAPTKTIC L +1owf_A +-1 --RTRT--RTNTPAQT-- L ! +2np2_A --3 --RT--RARANAPAKT-- L ! +1owf_B +-2 QTIT--RTRT--PT---- +4qju_B |
home
updated Tue Dec 19 06:00:26 2023
