| structure name | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE (PROTEIN CATABOLITE GENE ACTIVATOR PROTEIN CAP) |
| reference | Parkinson et al. Nat.Struct.Biol. 3 837 1996 |
| source | Escherichia coli |
| experiment | X-ray (resolution=2.70, R-factor=?) |
| structural superfamily | cAMP-binding domain-like;"Winged helix" DNA-binding domain; |
| sequence family | Cyclic nucleotidebinding domain; Bacterial regulatory proteins, crp famil; |
| multimeric complexes | 1cgp_AB 1lb2_ABE 3iyd_ACDFGH 3n4m_ABC 5ciz_AB 6pb4_CDFGH 6pb5_CFGH 6pb6_CDFGH |
| reference complex | 3n4m_B |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | QSRFTR |
Estimated binding specificities ?
readout + contact + contact | 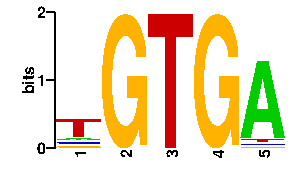 |
A | 12 0 0 0 85 C | 12 0 0 0 3 G | 12 96 0 96 3 T | 60 0 96 0 5scan! |
home
updated Mon Dec 18 05:58:49 2023
