Interface of input file ?
legend pdf

legend
| structure name | CRYO-EM STRUCTURE OF SPX-DEPENDENT TRANSCRIPTION ACTIVATION COMPLEX (RNA POLYMERASE SIGMA FACTOR SIGA) |
| source | BACILLUS SUBTILIS |
| experiment | NMR |
| structural superfamily | Sigma2 domain of RNA polymerase sigma factors;Sigma3 and sigma4 domains of RNA polymerase sigma factors; |
| sequence family | Sigma70 region 3; Sigma70 region 2; Sigma70, region 4; Sigma70 factor, region 12; HTH DNA binding domain; |
| redundant complexes | 7ckq |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | VRLEARVRRSTYTWRQTRIHEILFTERQ |
Estimated binding specificities ?
| contact | 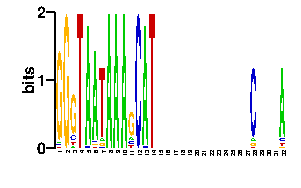 |
A | 7 24 24 24 7 24 24 24 24 24 24 24 24 24 24 24 24 24 90 0 0 7 0 0 96 7 90 96 0 7 0 2 C | 9 24 24 24 7 24 24 24 24 24 24 24 24 24 24 24 24 24 2 0 0 65 0 0 0 7 2 0 0 10 0 0 G | 7 24 24 24 75 24 24 24 24 24 24 24 24 24 24 24 24 24 2 0 96 10 0 0 0 7 2 0 0 69 96 90 T | 73 24 24 24 7 24 24 24 24 24 24 24 24 24 24 24 24 24 2 96 0 14 96 96 0 75 2 0 96 10 0 4scan! |
Dendrogram of similar interfaces ?
matrix formatVGRGLG----ETAT--RT--------VART------------RA------STTAYATAWA--RAQTTARGIA------HTEA------------IG--------LGFG--TT--ECRT--QT L +----7f75_F ! TAQALAITLTET--NTRTLT----------LA------FADA--RA--RTSCTAYATCWTWC--QCEARG------ITHGKC------------------------------RGET--RC-- L +----------------3 +6omf_F ! ! +-1 --------------NTRGLT----YA----FG------FAEA--RAFAKTSTTAYATCWTWTRCQTNARG----RTVGHTECIT----RARAATIGDGEG--SG--FGLTTTRGECRCRTQC L ! +---2 +4xln_F --5 ! ------------------LT----------------KTFAEA--RAFA--SATAYATA--WTRA------------------------------IG----DG----FG-------------- ! +6kqf_F ! --------------NTRTLT------------------FADA--------STTCYATTWTWG--QA--RG------------------------------------------RCEC----QT ! +-------------6wmr_Z +-------4 --------------NTRGLTIGKTYT------LTKT----EA----YAKTSATAYATAWGWTRCQATTRGITATRTVAHGET--NANA--RA------------------------------ +-------------6n4c_F |
home
updated Tue Dec 17 09:13:26 2024
