| structure name | CRYSTAL STRUCTURE OF THE BACTERIOPHAGE MU TRANSPOSOSOME |
| reference | Montano et al. Nature 491 413 2012 |
| source | ENTEROBACTERIA PHAGE MU |
| experiment | X-ray (resolution=3.71, R-factor=?) |
| structural superfamily | Ribonuclease H-like;Homeodomain-like;mu transposase, C-terminal domain; |
| reference complex | 4fcy_B |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RDYRR/STRDYRSRAF |
Estimated binding specificities ?
readout + contact + contact | 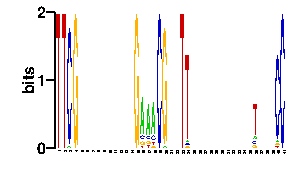 |
A | 0 3 24 24 24 61 24 24 24 24 24 24 24 24 24 24 24 84 96 24 24 3 0 24 3 3 3 24 24 24 24 24 24 24 24 24 24 0 0 96 96 C | 0 3 24 24 24 11 24 24 24 24 24 24 24 24 24 24 24 6 0 24 24 90 0 24 14 11 92 24 24 24 24 24 24 24 24 24 24 96 0 0 0 G | 96 87 24 24 24 13 24 24 24 24 24 24 24 24 24 24 24 3 0 24 24 3 96 24 7 13 0 24 24 24 24 24 24 24 24 24 24 0 96 0 0 T | 0 3 24 24 24 11 24 24 24 24 24 24 24 24 24 24 24 3 0 24 24 0 0 24 72 69 1 24 24 24 24 24 24 24 24 24 24 0 0 0 0scan! |
home
updated Fri Dec 15 18:52:58 2023
