| structure name | STRUCTURE OF THE EPSTEIN-BARR VIRUS ZEBRA PROTEIN (BZLF1 TRANS-ACTIVATOR PROTEIN) |
| reference | Artero et al. Mol.Cell 21 565 2006 |
| source | HUMAN HERPESVIRUS 4 |
| experiment | X-ray (resolution=2.25, R-factor=0.232) |
| structural superfamily | Leucine zipper domain; |
| sequence family | bZIP transcription factor; |
| multimeric complexes | 2c9l_YZ 2c9n_YZ 5szx_AB 7nx5_AB |
| redundant complexes |  2c9n_Y
5szx_B
7nx5_F 2c9n_Y
5szx_B
7nx5_F
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | NAASR |
Estimated binding specificities ?
| readout + contact | 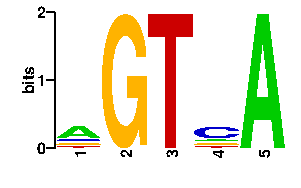 |
A | 0 16 96 0 13 C | 0 13 0 96 16 G | 0 54 0 0 13 T | 96 13 0 0 54scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: ZEBRA | ||
| TGAGCCA | PubMed | 2c9l_Z(6.10e-03), 2c9l_YZ(4.25e-07), 2c9n_YZ(4.09e-07), ... (total=5) |
| TGACATCA | PubMed | 2c9l_Z(1.77e-03) |
| TGAGTCA | PubMed | 2c9l_Z(7.17e-07), 2c9l_YZ(1.11e-12), 2c9n_YZ(1.05e-12), ... (total=5) |
| TGAGCAA | PubMed | 2c9l_YZ(1.66e-03), 2c9n_YZ(2.29e-03), 5szx_AB(1.05e-02), ... (total=4) |
Dendrogram of similar interfaces ?
matrix format----NC----ATAT----STRG-- L +-----2c9l_Z +-----------1 ------NCRT--ATAT----STRG +---2 +-----1t2k_D ! ! ----NC----AAAT------RT-- L +-3 +-----------------1a02_F ! ! NT----ATAT----STRG------ L --4 +---------------------1dh3_C ! --NC----AT------CTRA---- +-----------------------1s9k_E |
home
updated Tue Dec 19 06:23:54 2023
