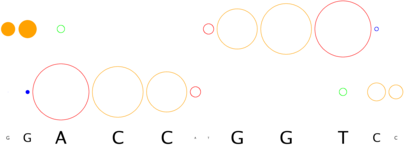These frequently asked questions will be updated with your
feedback, thanks.
-
I can't find PDB entry 1ABC, which I know is a transcription factor, why it is not in 3D-footprint?
This database considers only PDB structures that comprise both protein and DNA atomic coordinates, as only these structures
provide descriptions of the binding interface. Most likely your 1ABC entry includes only protein coordinates and therefore
is left out. However, if you find a missing entry that really should be included in the database please send feedback.
-
Why are there complexes with no interface graph?
In exceptional cases the 3D-footprint pipeline might fail to automatically recognize DNA duplexes in the original PDB coordinates.
In such cases it is still possible to generate an interface graph or footprint by editing the DNA coordinates of the original PDB file,
leaving only the relevant DNA chain, and submitting it to the
interactive 3D-footprint form.
Please check this sample file for a set of edited coordinates for sample complex 1fjl_C,
after leaving only DNA chain F.
-
How does 3D-footprint handle complexes with missing nucleotides or breaks in the DNA duplex?
Often the original molecular descriptions of proteins docked to DNA include breaks or molecular conformations that clearly are not helical.
3D-footprint attemps to handle all available complexes, but it considers only helical segments of DNA. For instance, the interface graph of
restriction enzyme Ecl18kI, captured in complex 2gb7_A, shows a break in
both DNA strands, as the original complex has a couple of flipped nucleotides, that were not represented in the graph as they are not in helical
conformation. Thus, it would appear that the structure-based binding consensus is CCGG, while the cognate restriction site of this enzyme
is actually CCnGG.
-
Why does 3D-footprint report monomers in complex with DNA?
We believe that taking monomers is the only way to possibly compare DNA-binding domains across superfamilies, preventing biases in the data,
as a vast majority of multimeric complexes contain redundant atomic interactions.
However, most often proteins recognize specific DNA sequences as dimers or higher order multimers, and these are linked as
multimeric complexes.
Users are advised to take specificities of multimeric complexes when available, such as entry
1cgp_AB for the global transcription factor
CRP in Escherichia coli (also accesible from entry 1zrf_A).
-
When I click on the structural superfamily link I get no matches, what does this mean?
The structural superfamily annotation of 3D-footprint entries is based on the Structural Classification of Proteins(SCOP) database, which is curated by experts and regularly updated. Recently published protein-DNA complexes usually have not yet been included in SCOP, and therefore the user gets this message. How is then that 3D-footprint labels these entries as members of superfamilies? We rely on the HMMER searches of the Superfamily library.
More generally, as 3D-footprint entries are linked to a variety of external resources, each with different release frequency, external links might fail to report
matches for recent entries.
-
Why does a DNA motif seem to be reversed?
DNA strands reported in PDB files are given in the usual 5'->3' orientation, which is indicated in interface graphs
with arrows as phosphodiester bonds. Consequently all DNA motifs reported in 3D-footprint are reported in this orientation. However, as there are two
complementary strands, it is not assured that the choosen strand will correspond to the preferred site description for each user.
-
Why is complex 1ABC_A a monomer when the paper reporting it describes a dimer?
Often molecular structures of multimeric complexes are deposited in the PDB as monomers that can be used to build a biologically relevant multimer
with help from symmetry matrices provided as REMARK 350 lines.
This is the case for the LEAFY transcription factor, described in
entry 2vy1_A, which is known to be a dimer. By taking the public
set of coordinates and applying the
matrix included there
REMARK 350 BIOMOLECULE: 1
REMARK 350 AUTHOR DETERMINED BIOLOGICAL UNIT: TETRAMERIC
REMARK 350 SOFTWARE DETERMINED QUATERNARY STRUCTURE: TETRAMERIC
REMARK 350 SOFTWARE USED: PISA
REMARK 350 APPLY THE FOLLOWING TO CHAINS: A, W
REMARK 350 BIOMT1 2 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 2 0.000000 -1.000000 0.000000 0.00000
REMARK 350 BIOMT3 2 0.000000 0.000000 -1.000000 0.00000
it is possible to build a dimeric complex, such as this one, that we can feed to the
interactive 3D-footprint form in order to produce this dimeric report:
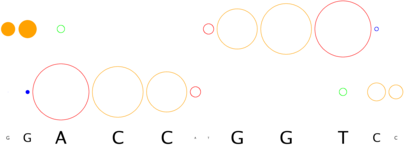
| readout + contact |  |
A | 6 2 62 0 13 41 11 5 1 12 40 36
C | 13 4 11 95 67 22 21 11 0 11 46 38
G | 41 45 12 0 9 24 24 67 94 11 5 11
T | 36 45 11 1 7 9 40 13 1 62 5 11
|
There are other examples like this in the PDB, but unfortunately building multimeric complexes cannot be safely handled automatically. Users are
therefore encouraged to manually build these complexes for interactive 3D-footprint analyses.
-
How can I get the PDB coordinates for any 3D-footprint entry?
The only set of coordinates that is currently distributed from our server is the compressed
non-redundant set of complexes in the download area.
In order to get the coordinates for any individual complex please follow the structure name link at the top, which
will take you to the corresponding prime entry in the Protein Data Bank, from where several download options are available.
-
Why is it that complex 1ABC_A is not included in file 'list_complexes_nr50_interface_nr70.txt'?
The file
list_complexes_nr50_interface_nr70.txt,
available in the downloads area,
contains a selection of non-redundant complexes according to two criteria: 50%ID
(protein sequence identity less than 50%) and 70%ID (less than 70% of interface identity). Complexes that fail any of those
criteria are not included in the list, as it's the case for 1ABC_A.
-
Why does my complex not produce a readout logo in 'interactive 3D-footprint'?
The format of your input DNA coordinates might be the reason for this, as the underlying DNAPROT algorithm
requires a well-defined DNA duplex in order to run. The fix should be just a quick edit of your input PDB file,
checking that only two DNA chains are included.
home