DNA Binding Motif
| Accessions: | 5t7x_AB (3D-footprint 20231221) |
| Names: | Epstein-Barr nuclear antigen 1 |
| Organisms: | Epstein-Barr virus (strain GD1) (HHV-4), Human herpesvirus 4 |
| Libraries: | 3D-footprint 20231221 1 1 Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic acids research 38:D91-7 (2010). [Pubmed] |
| Description: | Crystal structure of HHV-4 EBNA1 DNA binding domain (patient-derived, nasopharyngeal carcinoma) bound to DNA |
| Length: | 18 |
| Consensus: | GGATAGCnnntGcTACCC |
| Weblogo: | 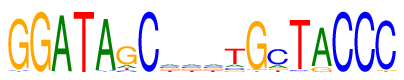 |
| PSSM: | P0 A C G T 01 0 2 92 2 G 02 0 0 96 0 G 03 94 0 2 0 A 04 0 0 0 96 T 05 92 2 0 2 A 06 12 7 68 9 G 07 0 96 0 0 C 08 24 24 24 24 n 09 24 24 24 24 n 10 24 24 24 24 n 11 12 11 9 64 t 12 1 2 91 2 G 13 14 64 9 9 c 14 0 3 2 91 T 15 83 0 12 1 A 16 0 96 0 0 C 17 0 96 0 0 C 18 0 91 3 2 C |
| Type: | Heterodimer |
| Binding TFs: | 5t7x_A (Epstein Barr virus nuclear antigen-1, DNA-binding domain) 5t7x_B (Epstein Barr virus nuclear antigen-1, DNA-binding domain) |
| Binding Sites: | 5t7x_C 5t7x_D |
| Publications: | Dheekollu J, Malecka K, Wiedmer A, Delecluse HJ, Chiang AK, Altieri DC, Messick TE, Lieberman PM. Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr Virus episomal latency. Oncotarget 8:7248-7264 (2017). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.