DNA Binding Motif
| Accessions: | CN0004.1 (JASPAR 2024) |
| Names: | LM4 |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Length: | 22 |
| Consensus: | gGcmTkCTGGGArwTGTAGTyy |
| Weblogo: | 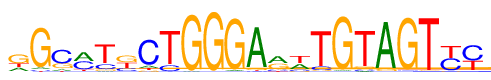 |
| PSSM: | P0 A C G T 01 392 83 960 442 g 02 91 64 1662 60 G 03 168 1150 444 115 c 04 850 767 91 169 m 05 77 361 153 1286 T 06 67 386 787 637 k 07 165 1451 117 144 C 08 36 293 18 1530 T 09 38 4 1822 13 G 10 13 8 1846 10 G 11 59 16 1780 22 G 12 1661 44 112 60 A 13 822 229 681 145 r 14 617 216 176 868 w 15 37 191 63 1586 T 16 52 22 1798 5 G 17 71 92 108 1606 T 18 1723 17 113 24 A 19 21 37 1790 29 G 20 15 29 46 1787 T 21 42 875 81 879 y 22 63 1080 68 666 y |
| Publications: | Xie X, Mikkelsen T.S, Gnirke A, Lindblad-Toh K, Kellis M, Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proceedings of the National Academy of Sciences of the United States of America 104:7145-50 (2007). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.