DNA Binding Motif
| Accessions: | RFX4_DBD_1 (HumanTF 1.0), MA0799.1 (JASPAR 2024) |
| Names: | RFX4 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF 1.0 1, JASPAR 2024 2 1 Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | Site type: dimeric; SELEX cycle: 3, HT-SELEX |
| Length: | 16 |
| Consensus: | cGTwrCYATGGywACs |
| Weblogo: | 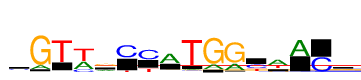 |
| PSSM: | P0 A C G T 01 2196 4157 2887 2408 c 02 2259 17 15889 246 G 03 417 380 203 13637 T 04 6538 2705 121 10463 w 05 4222 464 6675 2315 r 06 3 9323 18 1626 C 07 1 8836 45 3472 Y 08 10325 856 936 967 A 09 965 664 523 15458 T 10 2944 68 14990 1 G 11 4464 559 13778 107 G 12 1340 5757 98 3910 y 13 8911 127 1785 4986 w 14 9968 145 189 25 A 15 33 8784 11 304 C 16 2000 3460 5109 1853 s |
| Type: | Heterodimer |
| Binding TFs: | RFX4_DBD (RFX DNA-binding domain, Poxvirus D5 protein-like) Q33E94 (RFX DNA-binding domain) Q33E94 |
| Publications: | Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] Emery P., Strubin M., Hofmann K., Bucher P., Mach B., Reith W. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol. Cell. Biol. 16:4486-4494 (1996). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.