DNA Binding Motif
| Accessions: | MA1327.1 (JASPAR 2024), M0851 (AthalianaCistrome v4_May2016) |
| Names: | ATHB-23, ATHB23, ATHB23.DAP, T22039; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | JASPAR 2024 1, AthalianaCistrome v4_May2016 2 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] 2 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Notes: | DAP-seq |
| Length: | 22 |
| Consensus: | wwwhwTAATTARtwawwwawww |
| Weblogo: | 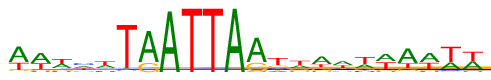 |
| PSSM: | P0 A C G T 01 364 22 33 180 w 02 318 43 52 186 w 03 201 64 94 240 w 04 164 167 117 151 h 05 183 89 105 222 w 06 26 16 13 544 T 07 455 1 143 0 A 08 593 0 0 6 A 09 0 0 0 599 T 10 0 0 0 599 T 11 599 0 0 0 A 12 423 19 155 2 R 13 73 101 123 302 t 14 157 62 146 234 w 15 300 46 124 129 a 16 227 104 89 179 w 17 156 71 78 294 w 18 252 37 67 243 w 19 373 21 71 134 a 20 294 44 50 211 w 21 165 32 31 371 w 22 235 33 47 284 w |
| Binding TFs: | T22039 (ZF-HD protein dimerisation region) |
| Binding Sites: | MA1327.1.1 MA1327.1.10 MA1327.1.11 MA1327.1.12 MA1327.1.13 MA1327.1.14 MA1327.1.15 MA1327.1.16 MA1327.1.17 MA1327.1.18 MA1327.1.19 MA1327.1.2 MA1327.1.20 MA1327.1.3 MA1327.1.4 MA1327.1.5 MA1327.1.6 MA1327.1.7 MA1327.1.8 MA1327.1.9 |
| Publications: | Johannesson H., Wang Y., Engstrom P. DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol. 45:63-73 (2001). [Pubmed] O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.