DNA Binding Motif
| Accessions: | MA1374.1 (JASPAR 2024), M0313 (AthalianaCistrome v4_May2016) |
| Names: | IDD7, IDD7.DAP, T02300; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | JASPAR 2024 1, AthalianaCistrome v4_May2016 2 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] 2 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Notes: | DAP-seq |
| Length: | 18 |
| Consensus: | mrsAAAAcGACAAAAawa |
| Weblogo: | 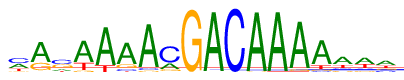 |
| PSSM: | P0 A C G T 01 190 232 44 119 m 02 394 20 153 18 r 03 95 224 197 69 s 04 400 3 36 146 A 05 489 1 0 95 A 06 424 49 88 24 A 07 528 31 23 3 A 08 136 375 57 17 c 09 2 0 583 0 G 10 575 9 0 1 A 11 0 585 0 0 C 12 574 0 11 0 A 13 576 2 6 1 A 14 540 11 29 5 A 15 441 25 39 80 A 16 293 89 66 137 a 17 267 77 62 179 w 18 244 95 108 138 a |
| Type: | Heterodimer |
| Binding TFs: | Q8H1F5 / T02300 (Zinc finger, C2H2 type, Zinc-finger double-stranded RNA-binding, C2H2-type zinc finger) Q8H1F5 |
| Binding Sites: | MA1374.1.1 MA1374.1.10 MA1374.1.11 MA1374.1.12 MA1374.1.13 MA1374.1.14 MA1374.1.15 MA1374.1.16 MA1374.1.17 MA1374.1.18 MA1374.1.19 MA1374.1.2 MA1374.1.20 MA1374.1.3 MA1374.1.4 MA1374.1.5 MA1374.1.6 MA1374.1.7 MA1374.1.8 MA1374.1.9 |
| Publications: | Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y. The arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet : (2013;9(9):e1003759.). [Pubmed] O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.