DNA Binding Motif
| Accessions: | MA1103.1 (JASPAR 2024) |
| Names: | FOXK2 |
| Organisms: | Homo sapiens |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | ChIP-seq |
| Length: | 11 |
| Consensus: | awgTAAACAar |
| Weblogo: | 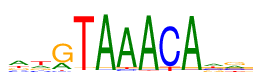 |
| PSSM: | P0 A C G T 01 3349 1353 1566 1709 a 02 2030 1315 1549 3083 w 03 1815 369 5407 386 g 04 116 53 140 7668 T 05 7730 119 62 66 A 06 7033 400 113 431 A 07 7698 75 57 147 A 08 91 6964 79 843 C 09 7680 67 113 117 A 10 2431 1875 1774 1897 a 11 2217 1882 2684 1194 r |
| Type: | Heterodimer |
| Binding TFs: | Q01167 (Fork head domain, FHA domain) Q01167 |
| Binding Sites: | MA1103.1.1 MA1103.1.10 / MA1103.1.5 MA1103.1.11 MA1103.1.12 MA1103.1.13 MA1103.1.14 MA1103.1.15 MA1103.1.16 MA1103.1.17 MA1103.1.18 MA1103.1.19 MA1103.1.2 MA1103.1.20 MA1103.1.3 MA1103.1.4 MA1103.1.6 MA1103.1.7 MA1103.1.8 MA1103.1.9 |
| Publications: | Chen X, Ji Z, Webber A, Sharrocks AD. Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors. Nucleic Acids Res 44:1566-78 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.