DNA Binding Motif
| Accessions: | MA1399.1 (JASPAR 2024), M0607 (AthalianaCistrome v4_May2016) |
| Names: | At5g08520, At5g08520.DAP, T26197; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | JASPAR 2024 1, AthalianaCistrome v4_May2016 2 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] 2 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Notes: | DAP-seq |
| Length: | 13 |
| Consensus: | dwwdwrGATAAGr |
| Weblogo: | 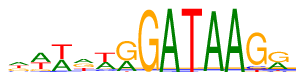 |
| PSSM: | P0 A C G T 01 194 69 166 169 d 02 292 23 72 211 w 03 248 17 15 318 w 04 209 42 180 167 d 05 221 59 30 288 w 06 197 2 376 23 r 07 0 0 596 2 G 08 598 0 0 0 A 09 0 0 0 598 T 10 590 0 8 0 A 11 598 0 0 0 A 12 5 53 495 45 G 13 204 40 312 42 r |
| Type: | Heterodimer |
| Binding TFs: | Q9FNN6 / T26197 (Myb-like DNA-binding domain) Q9FNN6 |
| Binding Sites: | MA1399.1.1 MA1399.1.10 MA1399.1.11 MA1399.1.12 MA1399.1.13 MA1399.1.14 MA1399.1.15 MA1399.1.16 MA1399.1.17 MA1399.1.18 MA1399.1.19 MA1399.1.2 MA1399.1.20 MA1399.1.3 MA1399.1.4 MA1399.1.5 MA1399.1.6 MA1399.1.7 MA1399.1.8 MA1399.1.9 |
| Publications: | Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014 Sep 11;158(6):1431-43. doi: 10.1016/j.cell.2014.08.009. [Pubmed] O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.