DNA Binding Motif
| Accessions: | M0482 (AthalianaCistrome v4_May2016) |
| Names: | AGL15.ampDAP, T23985; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | AthalianaCistrome v4_May2016 1 1 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Length: | 21 |
| Consensus: | wwcTTTCYwtWTTkGGwAAst |
| Weblogo: | 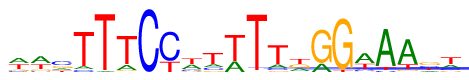 |
| PSSM: | P0 A C G T 01 225 64 76 219 w 02 262 46 82 194 w 03 117 235 139 93 c 04 49 38 6 491 T 05 16 0 2 566 T 06 89 11 27 457 T 07 1 583 0 0 C 08 0 427 0 157 Y 09 197 85 57 245 w 10 101 91 48 344 t 11 172 3 1 408 W 12 7 10 1 566 T 13 87 54 34 409 T 14 135 40 174 235 k 15 122 1 461 0 G 16 0 5 552 27 G 17 309 62 37 176 w 18 505 24 7 48 A 19 440 12 40 92 A 20 94 160 200 130 s 21 145 85 70 284 t |
| Binding TFs: | T23985 (SRF-type transcription factor (DNA-binding and dimerisation domain), K-box region) |
| Publications: | O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.