DNA Binding Motif
| Accessions: | 6edb_AB (3D-footprint 20231221) |
| Names: | Sex-determining region Y protein,Cyclic GMP-AMP synthase |
| Organisms: | Homo sapiens |
| Libraries: | 3D-footprint 20231221 1 1 Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic acids research 38:D91-7 (2010). [Pubmed] |
| Description: | Crystal structure of SRY.hcGAS-21bp dsDNA complex |
| Length: | 14 |
| Consensus: | cATTgkknnnnnCc |
| Weblogo: | 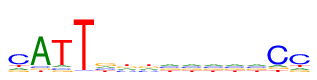 |
| PSSM: | P0 A C G T 01 13 56 13 14 c 02 78 6 6 6 A 03 7 7 7 75 T 04 1 2 1 92 T 05 20 20 36 20 g 06 22 22 25 27 k 07 22 22 25 27 k 08 24 24 24 24 n 09 24 24 24 24 n 10 24 24 24 24 n 11 24 24 24 24 n 12 24 24 24 24 n 13 9 69 9 9 C 14 13 56 13 14 c |
| Binding TFs: | 6edb_B (HMG (high mobility group) box, Domain of unknown function (DUF1898)) |
| Binding Sites: | 6edb_C 6edb_D |
| Publications: | Xie W, Lama L, Adura C, Tomita D, Glickman JF, Tuschl T, Patel DJ. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc Natl Acad Sci U S A 116:11946-11955 (2019). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.