DNA Binding Motif
| Accessions: | TBX21_full_2 (HumanTF 1.0), MA0690.1 (JASPAR 2024) |
| Names: | TBX21 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF 1.0 1, JASPAR 2024 2 1 Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | Site type: monomeric; SELEX cycle: 3, HT-SELEX |
| Length: | 10 |
| Consensus: | aAGGTGTGAA |
| Weblogo: | 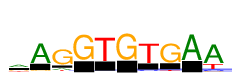 |
| PSSM: | P0 A C G T 01 2322 263 937 872 a 02 4395 30 359 146 A 03 612 239 4395 398 G 04 2 17 4395 33 G 05 6 180 16 4395 T 06 1 0 4395 2 G 07 185 331 34 4395 T 08 80 126 4395 364 G 09 4395 0 38 0 A 10 4395 441 317 1287 A |
| Type: | Heterodimer |
| Binding TFs: | TBX21_full (T-box) Q9UL17 (T-box) Q9UL17 |
| Publications: | Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet : (2005;39:219-39.). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.