DNA Binding Motif
| Accessions: | MA1258.1 (JASPAR 2024), M0084 (AthalianaCistrome v4_May2016) |
| Names: | DREB2, DREB2A, DREB2.DAP, T24768; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | JASPAR 2024 1, AthalianaCistrome v4_May2016 2 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] 2 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Notes: | DAP-seq |
| Length: | 21 |
| Consensus: | wgrydrwGkCGGTGgwgrwrr |
| Weblogo: | 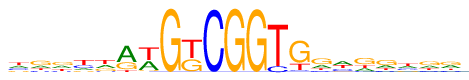 |
| PSSM: | P0 A C G T 01 166 130 102 183 w 02 138 82 243 118 g 03 155 78 204 144 r 04 103 149 76 253 y 05 166 54 146 215 d 06 360 9 162 50 r 07 219 19 35 308 w 08 0 0 581 0 G 09 18 0 226 337 k 10 2 579 0 0 C 11 0 0 581 0 G 12 1 0 580 0 G 13 0 78 0 503 T 14 16 25 413 127 G 15 145 68 267 101 g 16 212 129 67 173 w 17 127 48 270 136 g 18 149 74 258 100 r 19 160 129 108 184 w 20 149 87 225 120 r 21 182 91 206 102 r |
| Binding TFs: | T24768 (AP2 domain) |
| Publications: | Medina J, Bargues M, Terol J, Pérez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119:463-70 (1999). [Pubmed] O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.