DNA Binding Motif
| Accessions: | 6pst_L (3D-footprint 20231221) |
| Names: | RNA polymerase sigma factor RpoD |
| Organisms: | Escherichia coli, strain K12 |
| Libraries: | 3D-footprint 20231221 1 1 Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic acids research 38:D91-7 (2010). [Pubmed] |
| Description: | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5b) with TraR and mutant rpsT P2 promoter |
| Length: | 30 |
| Consensus: | GATGCGCCnnTnnnnnnnnnnnnnGTCAAT |
| Weblogo: | 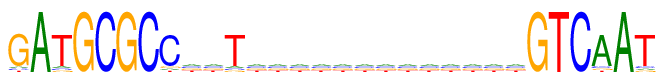 |
| PSSM: | P0 A C G T 01 5 5 81 5 G 02 96 0 0 0 A 03 6 5 5 80 T 04 0 0 96 0 G 05 0 96 0 0 C 06 0 0 96 0 G 07 0 96 0 0 C 08 5 80 6 5 C 09 24 24 24 24 n 10 24 24 24 24 n 11 6 5 5 80 T 12 24 24 24 24 n 13 24 24 24 24 n 14 24 24 24 24 n 15 24 24 24 24 n 16 24 24 24 24 n 17 24 24 24 24 n 18 24 24 24 24 n 19 24 24 24 24 n 20 24 24 24 24 n 21 24 24 24 24 n 22 24 24 24 24 n 23 24 24 24 24 n 24 24 24 24 24 n 25 0 0 96 0 G 26 0 0 0 96 T 27 0 96 0 0 C 28 80 6 5 5 A 29 96 0 0 0 A 30 5 6 5 80 T |
| Binding TFs: | 6pst_L (Sigma-70 factor, region 1.2, Sigma-70 factor, region 1.1, Sigma-70 region 3, Sigma-70 region 2 , Sigma-70, region 4, Sigma-70, non-essential region) |
| Binding Sites: | 6pst_O 6pst_P |
| Publications: | Chen J, Chiu C, Gopalkrishnan S, Chen AY, Olinares PDB, Saecker RM, Winkelman JT, Maloney MF, Chait BT, Ross W, Gourse RL, Campbell EA, Darst SA. Stepwise Promoter Melting by Bacterial RNA Polymerase. Mol Cell 78:275-288 (2020). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.