DNA Binding Motif
| Accessions: | MA1247.1 (JASPAR 2024), M0100 (AthalianaCistrome v4_May2016) |
| Names: | AT1G28160, ERF087, AT1G28160.DAP, T06007; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | JASPAR 2024 1, AthalianaCistrome v4_May2016 2 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] 2 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Notes: | DAP-seq |
| Length: | 15 |
| Consensus: | ygrCGGCGGmGrhgg |
| Weblogo: | 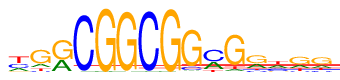 |
| PSSM: | P0 A C G T 01 129 178 28 260 y 02 97 38 374 86 g 03 212 11 366 6 r 04 8 577 0 10 C 05 0 0 595 0 G 06 6 1 586 2 G 07 0 586 0 9 C 08 0 0 594 1 G 09 15 55 511 14 G 10 200 314 11 70 m 11 42 15 485 53 G 12 165 64 301 65 r 13 182 165 65 183 h 14 133 42 294 126 g 15 125 69 296 105 g |
| Type: | Heterodimer |
| Binding TFs: | Q9FZ90 / T06007 (AP2 domain) Q9FZ90 |
| Binding Sites: | MA1247.1.2 MA1247.1.7 MA1247.1.13 / MA1247.1.9 MA1247.1.17 MA1247.1.10 MA1247.1.19 MA1247.1.18 MA1247.1.4 MA1247.1.5 MA1247.1.3 / MA1247.1.6 MA1247.1.1 MA1247.1.14 MA1247.1.11 MA1247.1.12 MA1247.1.8 MA1247.1.20 MA1247.1.15 MA1247.1.16 |
| Publications: | Hao D., Ohme-Takagi M., Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273:26857-26861 (1998). [Pubmed] O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.