DNA Binding Motif
| Accessions: | MA0390.1 (JASPAR 2024) |
| Names: | STB3 |
| Organisms: | Saccharomyces cerevisiae |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | PBM |
| Length: | 21 |
| Consensus: | gtyyaaAwTTTTTCActyhkk |
| Weblogo: | 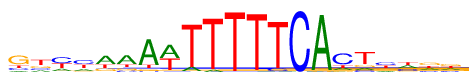 |
| PSSM: | P0 A C G T 01 98 168 546 185 g 02 166 173 202 457 t 03 182 415 57 343 y 04 194 379 107 318 y 05 507 59 189 243 a 06 547 120 151 181 a 07 776 35 71 116 A 08 499 27 33 440 w 09 99 11 2 886 T 10 53 5 1 939 T 11 10 4 2 982 T 12 13 0 15 970 T 13 0 64 20 915 T 14 3 990 2 4 C 15 965 7 25 2 A 16 150 587 84 177 c 17 129 120 137 613 t 18 166 326 200 306 y 19 308 253 125 312 h 20 233 170 315 280 k 21 226 182 296 293 k |
| Type: | Heterodimer |
| Binding TFs: | Q12427 (Putative Sin3 binding protein) Q12427 |
| Publications: | Newburger D.E, Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic acids research 37:D77-82 (2009). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.