DNA Binding Motif
| Accessions: | MA0495.1 (JASPAR 2024) |
| Names: | MAFF |
| Organisms: | Homo sapiens |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | ChIP-seq |
| Length: | 18 |
| Consensus: | gctrasTCAGCAwwwwtt |
| Weblogo: | 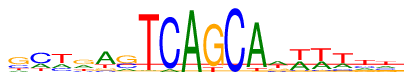 |
| PSSM: | P0 A C G T 01 6531 12389 26849 7989 g 02 13157 29577 2157 8867 c 03 12632 10615 2570 27941 t 04 14736 6027 21695 11300 r 05 32728 4789 5877 10364 a 06 8513 16391 25722 3132 s 07 998 322 313 52125 T 08 3238 50520 0 0 C 09 53327 0 0 431 A 10 387 0 47337 6034 G 11 0 53701 0 57 C 12 48740 381 746 3891 A 13 20032 7624 9364 16738 w 14 21358 3349 2603 26448 w 15 18219 4014 1898 29627 w 16 14137 7157 3512 28952 w 17 10417 11010 8426 23905 t 18 12741 10791 9498 20728 t |
| Type: | Heterodimer |
| Binding TFs: | Q9ULX9 (bZIP Maf transcription factor) Q9ULX9 |
| Binding Sites: | MA0495.1.1 MA0495.1.10 MA0495.1.11 MA0495.1.12 MA0495.1.13 MA0495.1.14 MA0495.1.15 MA0495.1.16 MA0495.1.17 MA0495.1.18 MA0495.1.19 MA0495.1.2 MA0495.1.20 MA0495.1.3 MA0495.1.4 MA0495.1.5 MA0495.1.6 MA0495.1.7 MA0495.1.8 MA0495.1.9 |
| Publications: | Kataoka K., Noda M., Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14:700-712 (1994). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.