DNA Binding Motif
| Accessions: | MA1113.1 (JASPAR 2024) |
| Names: | PBX2 |
| Organisms: | Homo sapiens |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | ChIP-seq |
| Length: | 12 |
| Consensus: | gtGAtTGACAgs |
| Weblogo: | 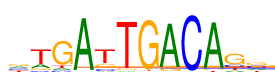 |
| PSSM: | P0 A C G T 01 607 604 708 576 g 02 449 176 266 1604 t 03 191 166 1897 241 G 04 2379 29 46 41 A 05 135 309 507 1544 t 06 57 22 117 2299 T 07 32 44 2350 69 G 08 2242 21 200 32 A 09 19 2419 25 32 C 10 2293 25 92 85 A 11 262 399 1456 378 g 12 512 754 793 436 s |
| Type: | Heterodimer |
| Binding TFs: | P40425 (Homeobox domain, PBC domain, Homeobox KN domain) P40425 |
| Binding Sites: | MA1113.1.1 MA1113.1.10 MA1113.1.11 MA1113.1.12 MA1113.1.13 MA1113.1.14 MA1113.1.15 MA1113.1.16 MA1113.1.17 MA1113.1.18 MA1113.1.19 MA1113.1.2 MA1113.1.20 MA1113.1.3 MA1113.1.4 MA1113.1.5 MA1113.1.6 MA1113.1.7 MA1113.1.8 MA1113.1.9 |
| Publications: | Shen W. F., Rozenfeld S., Kwong A., Kom ves L. G., Lawrence H. J., Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells.. Mol. Cell. Biol. 19:3051-3061 (1999). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.