DNA Binding Motif
| Accessions: | MA0378.1 (JASPAR 2024) |
| Names: | SFP1 |
| Organisms: | Saccharomyces cerevisiae |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | PBM |
| Length: | 21 |
| Consensus: | wywrdrAAAAwTTTTyywykg |
| Weblogo: | 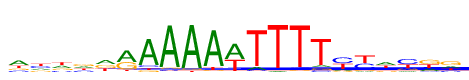 |
| PSSM: | P0 A C G T 01 427 159 161 250 w 02 202 293 194 309 y 03 289 174 192 343 w 04 250 217 293 238 r 05 353 77 297 271 d 06 533 42 342 82 r 07 782 73 71 73 A 08 898 36 46 18 A 09 904 9 50 35 A 10 855 14 42 86 A 11 541 34 16 408 w 12 86 42 14 855 T 13 35 50 9 904 T 14 18 46 36 898 T 15 73 71 73 782 T 16 126 405 91 376 y 17 82 354 103 459 y 18 327 201 165 306 w 19 115 428 154 301 y 20 217 83 374 324 k 21 216 169 405 208 g |
| Type: | Heterodimer |
| Binding TFs: | P32432 P32432 |
| Publications: | Newburger D.E, Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic acids research 37:D77-82 (2009). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.