DNA Binding Motif
| Accessions: | Tp73_DBD (HumanTF 1.0), MA0861.1 (JASPAR 2024) |
| Names: | Tp73 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF 1.0 1, JASPAR 2024 2 1 Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] 2 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | Site type: dimeric; SELEX cycle: 4, HT-SELEX |
| Length: | 18 |
| Consensus: | drCATGTcyrrACAyGym |
| Weblogo: | 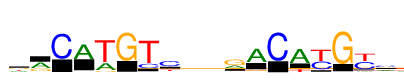 |
| PSSM: | P0 A C G T 01 3617 313 3891 3689 d 02 6206 429 3304 208 r 03 0 10829 0 0 C 04 8760 124 557 198 A 05 3837 293 806 11480 T 06 8 28 11787 1 G 07 130 2591 91 11892 T 08 1377 8161 783 2749 c 09 2166 2940 2230 4242 y 10 3488 2418 3558 2457 r 11 3988 441 7797 1002 r 12 9639 145 1414 442 A 13 0 11703 0 0 C 14 10125 143 279 2302 A 15 256 5965 130 8476 y 16 16 170 13026 39 G 17 993 6944 181 6995 y 18 4008 5125 519 2232 m |
| Type: | Heterodimer |
| Binding TFs: | O15350 (P53 DNA-binding domain, SAM domain (Sterile alpha motif), P53 tetramerisation motif) O15350 |
| Publications: | Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] Ethayathulla AS, Nguyen HT, Viadiu H. Crystal structures of the DNA-binding domain tetramer of the p53 tumor suppressor family member p73 bound to different full-site response elements. J Biol Chem 288:4744-54 (2013). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.