DNA Binding Motif
| Accessions: | BARX1_DBD_1 (HumanTF 1.0) |
| Names: | BARX1 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF 1.0 1 1 Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] |
| Notes: | Site type: dimeric; SELEX cycle: 3 |
| Length: | 17 |
| Consensus: | cvATTAAawavCrATTA |
| Weblogo: | 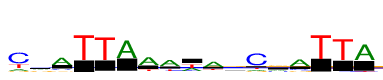 |
| PSSM: | P0 A C G T 01 420 1863 245 735 c 02 734 696 540 186 v 03 1696 305 235 134 A 04 7 9 4 2200 T 05 7 66 6 2166 T 06 1841 8 16 71 A 07 1720 115 148 416 A 08 1668 194 165 572 a 09 885 67 102 1033 w 10 1662 189 297 454 a 11 587 619 559 395 v 12 290 2124 259 293 C 13 628 520 1026 74 r 14 1716 396 191 213 A 15 4 4 0 2101 T 16 24 74 5 2097 T 17 1860 13 36 157 A |
| Binding TFs: | BARX1_DBD (Homeobox domain) |
| Publications: | Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, Palin K, Vaquerizas JM, Vincentelli R, Luscombe NM, Hughes TR, Lemaire P, Ukkonen E, Kivioja T, Taipale J. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013 Jan 17;152(1-2):327-39. [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.