DNA Binding Motif
| Accessions: | CUX1_RFX5_1 (HumanTF2 1.0) |
| Names: | CUX1_RFX5 |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 23 |
| Consensus: | rATCrATagsmdtrCwTaGyAAC |
| Weblogo: | 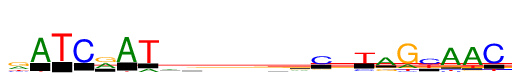 |
| PSSM: | P0 A C G T 01 360 148 510 173 r 02 870 2 8 40 A 03 2 2 1 870 T 04 21 870 2 35 C 05 423 12 447 36 r 06 870 0 4 40 A 07 145 0 0 870 T 08 383 171 165 150 a 09 207 121 365 177 g 10 214 232 270 154 s 11 270 285 115 199 m 12 219 123 285 242 d 13 160 210 137 363 t 14 306 135 344 85 r 15 64 648 83 76 C 16 320 184 115 250 w 17 33 147 24 666 T 18 658 77 212 37 a 19 179 56 870 111 G 20 51 518 18 352 y 21 870 58 107 86 A 22 870 37 63 153 A 23 24 870 19 107 C |
| Type: | Heterodimer |
| Binding TFs: | CUX1_TF1 (Homeobox domain, CUT domain, Homeobox KN domain) RFX5_TF2 (RFX DNA-binding domain, Poxvirus D5 protein-like) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.