DNA Binding Motif
| Accessions: | M0715 (AthalianaCistrome v4_May2016) |
| Names: | AGL95.DAP, T07153; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | AthalianaCistrome v4_May2016 1 1 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Length: | 15 |
| Consensus: | aGAAgcTTCTAGAAg |
| Weblogo: | 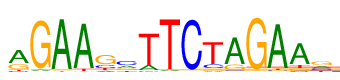 |
| PSSM: | P0 A C G T 01 359 42 147 47 a 02 22 7 556 10 G 03 562 6 15 12 A 04 542 4 13 36 A 05 61 116 331 87 g 06 145 241 128 81 c 07 84 20 9 482 T 08 22 3 10 560 T 09 8 562 11 14 C 10 30 84 44 437 T 11 454 23 105 13 A 12 8 9 568 10 G 13 543 15 13 24 A 14 431 28 62 74 A 15 136 130 234 95 g |
| Binding TFs: | T07153 |
| Publications: | O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.