DNA Binding Motif
| Accessions: | CUX1_SRF (HumanTF2 1.0) |
| Names: | CUX1_SRF |
| Organisms: | Homo sapiens |
| Libraries: | HumanTF2 1.0 1 1 Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
| Notes: | Site type: heterodimeric; Experiment type: CAP-SELEX |
| Length: | 23 |
| Consensus: | rCCwTAyAwGGtmrkrATCrATw |
| Weblogo: | 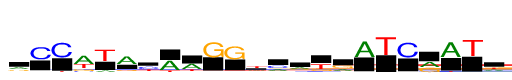 |
| PSSM: | P0 A C G T 01 1676 598 1024 487 r 02 242 3785 168 143 C 03 12 3785 0 375 C 04 3785 534 1144 1943 w 05 660 131 46 3785 T 06 3785 324 712 343 A 07 584 2066 173 1719 y 08 3118 40 96 666 A 09 2437 178 207 963 w 10 232 30 3785 8 G 11 128 108 3785 228 G 12 1180 840 428 2605 t 13 1303 1725 562 195 m 14 1656 531 980 618 r 15 171 180 1162 2271 k 16 1225 142 1885 533 r 17 3785 8 78 135 A 18 5 4 16 3785 T 19 43 3785 18 246 C 20 2001 45 1783 7 r 21 3785 6 5 369 A 22 166 9 2 3785 T 23 1340 370 691 1383 w |
| Type: | Heterodimer |
| Binding TFs: | CUX1_TF1 (Homeobox domain, CUT domain, Homeobox KN domain) SRF_TF1 / SRF_TF2 (SRF-type transcription factor (DNA-binding and dimerisation domain)) |
| Publications: | Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527:384-8 (2015). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.