DNA Binding Motif
| Accessions: | GlnR (DBTBS 1.0) |
| Names: | GlnR |
| Organisms: | Bacillus subtilis |
| Libraries: | DBTBS 1.0 1 1 Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic acids research 36:D93-6 (2008). [Pubmed] |
| Length: | 17 |
| Consensus: | TGTcAcrAAAwcTwACA |
| Weblogo: | 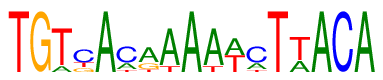 |
| PSSM: | P0 A C G T 01 0 0 0 1 T 02 0 0 1 0 G 03 0.15 0 0 0.85 T 04 0 0.58 0.19 0.22 c 05 1 0 0 0 A 06 0.25 0.65 0 0.11 c 07 0.61 0 0.26 0.13 r 08 0.85 0 0 0.15 A 09 1 0 0 0 A 10 0.85 0 0 0.15 A 11 0.50 0 0 0.50 w 12 0.25 0.65 0 0.11 c 13 0 0 0 1 T 14 0.33 0 0 0.67 w 15 1 0 0 0 A 16 0 1 0 0 C 17 1 0 0 0 A |
| Binding TFs: | GlnR (MerR family regulatory protein, MerR HTH family regulatory protein) |
| Binding Sites: | glnR_1 glnR_2 nasA_1 nasB_1 ureA_1 ureA_2 |
| Publications: | Gutowski J.C, Schreier H.J. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter sequences in vivo. Journal of bacteriology 174:671-81 (1992). [Pubmed] Nakano M.M, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. Journal of bacteriology 177:573-9 (1995). [Pubmed] Wray L.V, Ferson A.E, Fisher S.H. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. Journal of bacteriology 179:5494-501 (1997). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.