DNA Binding Motif
| Accessions: | M0668 (AthalianaCistrome v4_May2016) |
| Names: | ANAC045.ampDAP, T13400; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | AthalianaCistrome v4_May2016 1 1 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Length: | 16 |
| Consensus: | CkTGyrgwasAAGyAA |
| Weblogo: | 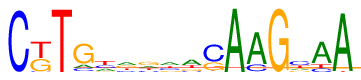 |
| PSSM: | P0 A C G T 01 4 586 0 0 C 02 0 23 246 321 k 03 0 2 0 588 T 04 81 71 419 19 G 05 144 155 64 227 y 06 190 93 182 125 r 07 142 99 218 131 g 08 197 145 99 149 w 09 263 68 112 147 a 10 20 369 176 25 s 11 590 0 0 0 A 12 468 107 15 0 A 13 0 0 590 0 G 14 102 234 93 161 y 15 420 81 6 83 A 16 576 0 0 14 A |
| Binding TFs: | T13400 (No apical meristem (NAM) protein) |
| Publications: | O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.