DNA Binding Motif
| Accessions: | MA0314.1 (JASPAR 2024) |
| Names: | HAP3 |
| Organisms: | Saccharomyces cerevisiae |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | COMPILED |
| Length: | 15 |
| Consensus: | tcTsATTGGyyvrra |
| Weblogo: | 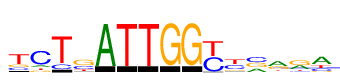 |
| PSSM: | P0 A C G T 01 0 516 537 1779 t 02 370 2074 298 326 c 03 0 461 0 2379 T 04 98 897 1473 484 s 05 2737 0 0 0 A 06 0 0 0 2737 T 07 0 0 0 2737 T 08 0 0 2737 0 G 09 0 0 2737 0 G 10 0 1480 0 1549 y 11 97 1169 405 1258 y 12 809 1175 1038 98 v 13 1458 265 918 339 r 14 1087 266 1351 339 r 15 1582 602 296 665 a |
| Type: | Heterodimer |
| Binding TFs: | P13434 (Core histone H2A/H2B/H3/H4, Histone-like transcription factor (CBF/NF-Y) and archaeal histone) P13434 |
| Publications: | Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic acids research 35:D127-31 (2007). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.