DNA Binding Motif
| Accessions: | MA0269.1 (JASPAR 2024) |
| Names: | AFT1 |
| Organisms: | Saccharomyces cerevisiae |
| Libraries: | JASPAR 2024 1 1 Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, Lemma RB, Lucas J, Cheneby J, Baranasic D, Khan A, Fornes O, Gundersen S, Johansen M, Hovig E, Lenhard B, Sandelin A, Wasserman WW, Parcy F, Mathelier A. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res : (2023). [Pubmed] |
| Notes: | PBM |
| Length: | 21 |
| Consensus: | ttacatAkTGCACCCsmwwkg |
| Weblogo: | 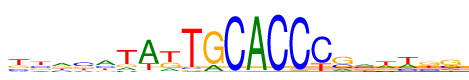 |
| PSSM: | P0 A C G T 01 106 205 231 456 t 02 155 249 220 374 t 03 362 197 221 218 a 04 179 396 222 200 c 05 426 231 154 187 a 06 136 78 142 641 t 07 716 25 31 226 A 08 145 31 288 535 k 09 34 47 39 878 T 10 160 10 812 16 G 11 10 966 11 10 C 12 961 9 15 14 A 13 8 972 9 10 C 14 25 955 2 16 C 15 19 740 13 225 C 16 181 257 494 65 s 17 265 252 241 240 m 18 287 155 183 373 w 19 276 180 68 474 w 20 123 225 357 292 k 21 154 236 458 150 g |
| Type: | Heterodimer |
| Binding TFs: | P22149 (Transcription factor AFT) P22149 |
| Publications: | Newburger D.E, Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic acids research 37:D77-82 (2009). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.