DNA Binding Motif
| Accessions: | M0678 (AthalianaCistrome v4_May2016) |
| Names: | ANAC046.DAP, T13495; |
| Organisms: | Arabidopsis thaliana |
| Libraries: | AthalianaCistrome v4_May2016 1 1 O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
| Length: | 21 |
| Consensus: | TwrCKTGtrrmACAmGyaAhc |
| Weblogo: | 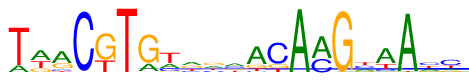 |
| PSSM: | P0 A C G T 01 91 2 1 482 T 02 271 1 112 192 w 03 331 58 151 36 r 04 9 567 0 0 C 05 0 0 398 178 K 06 0 0 0 576 T 07 119 15 439 3 G 08 134 105 22 315 t 09 201 86 165 124 r 10 158 106 177 135 r 11 187 148 125 116 m 12 393 26 86 71 A 13 5 407 89 75 C 14 575 0 1 0 A 15 353 221 1 1 m 16 1 0 571 4 G 17 142 159 83 192 y 18 358 68 16 134 a 19 562 0 0 14 A 20 147 180 92 157 h 21 96 262 85 133 c |
| Binding TFs: | T13495 (No apical meristem (NAM) protein) |
| Publications: | O'Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 165:1280-92 (2016). [Pubmed] |
Disclaimer and license
These data are available AS IS and at your own risk. The EEAD/CSIC do not give any representation or warranty nor assume any liability or responsibility for the data nor the results posted (whether as to their accuracy, completeness, quality or otherwise). Access to these data is available free of charge for ordinary use in the course of research. Downloaded data have CC-BY-NC-SA license. FootprintDB is also available at RSAT::Plants, part of the INB/ELIXIR-ES resources portfolio.