| structure name | CRYSTAL STRUCTURE OF THE FORKHEAD DOMAIN OF HUMAN FOXN1 IN COMPLEX WITH DNA (Forkhead box protein N1) |
| reference | Structural Genomics Consortium (SGC) |
| source | Homo sapiens |
| experiment | X-ray (resolution=1.61, R-factor=0.196) |
| structural superfamily | "Winged helix" DNA-binding domain; |
| sequence family | Fork head domain; |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | NSRHS |
Estimated binding specificities ?
| readout + contact | 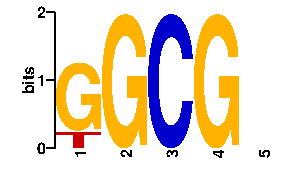 |
A | 19 76 0 0 0 0 C | 19 0 96 0 96 96 G | 19 20 0 96 0 0 T | 39 0 0 0 0 0scan! |
Dendrogram of similar interfaces ?
matrix format------------------------NAST--RGHT----STLT------------ L +--------7cby_C +------------------2 ----------------------------------------------------NG ! +--------7k63_U ! ----KAFA------------------------------------YANAKTFGMC ! +-----------3aaf_B ! +-3 ----------LT--------RT--YTPT----------------------RT-- L ! +-----4 +-----------6qil_A +-9 ! ! ------------------------NAST--RGHTNT--STLT------------ L ! ! +---6 +-------------7fj2_B ! ! ! ! --------RC----------------------------------------SG-- ! ! +--7 +-------------------7cow_T ! ! ! ! --------------ST--PTTC--STQC------RA------------------ L ! ! ! ! +---------------1ddn_B +-10 +-8 +-------5 ------------------TTHT----KG-------------------------- L ! ! ! +---------------7b0c_A ! ! ! ------------------------NAST--RTHT----ST-------------- ! ! +--------------------------3co7_C +-11 ! --------------------QTNAST--RGHTNT--STLA-------------- L ! ! ! +------2hdc_A ! ! +--------------------1 RT--------LT--NTHGTA----YTRG-------------------------- +-12 ! +------2efw_A ! ! ! --RC-------------------------------------------------- ! ! +----------------------------6l9z_S +-13 ! ------------RAST--ECTT--RAKA-------------------------- ! ! +----------------------------7oz3_B +-14 ! ------------------KGTA----TA------RG------------------ L ! ! +-----------------------------1xsd_A -15 ! --------------------SA-------------------------------- ! +-----------------------------2xcr_U ! --------DT----------RGRCTT----KT----------RT---------- L +-----------------------------2o9l_A |
home
updated Tue Dec 19 12:45:05 2023
