| structure name | ATP-DEPENDENT TYPE ISP RESTRICTION-MODIFICATION ENZYME LLABIII BOUND TO DNA (LLABIII) |
| reference | Saikrishnan et al. Nat.Chem.Biol. 11 870 2015 |
| source | LACTOCOCCUS LACTIS SUBSP. CREMORIS |
| experiment | X-ray (resolution=2.70, R-factor=0.221) |
| structural superfamily | S-adenosyl-L-methionine-dependent methyltransferases;P-loop containing nucleoside triphosphate hydrolases;Restriction endonuclease-like; |
| sequence family | Type III restriction enzyme, res subunit; Helicase conserved Cterminal domain; |
| multimeric complexes | 4xqk_AB |
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | KVNPYKKTNNIRKFIMKRDRYN |
Estimated binding specificities ?
| contact | 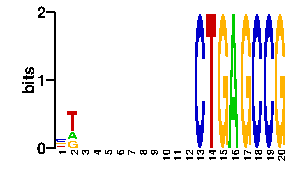 |
A | 21 21 24 24 24 24 24 24 24 24 24 24 0 0 0 96 0 0 0 0 C | 42 0 24 24 24 24 24 24 24 24 24 24 96 0 0 0 0 96 96 0 G | 10 21 24 24 24 24 24 24 24 24 24 24 0 0 96 0 96 0 0 96 T | 23 54 24 24 24 24 24 24 24 24 24 24 0 96 0 0 0 0 0 0scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: LLABIII | ||
| TNAGCC | PubMed | 4xqk_A(7.69e-04), 4xqk_AB(7.69e-04) |
home
updated Tue Dec 19 09:42:06 2023
