| structure name | X-RAY STRUCTURE OF ARCHAEAL CLASS II CPD PHOTOLYASE FROM METHANOSARCINA MAZEI IN COMPLEX WITH INTACT CPD-LESION (DEOXYRIBODIPYRIMIDINE PHOTOLYASE) |
| reference | Geisselbrecht et al. 'Embo 30 4437 2011 |
| source | METHANOSARCINA MAZEI |
| experiment | X-ray (resolution=2.20, R-factor=0.171) |
| structural superfamily | Cryptochrome/photolyase FAD-binding domain;Cryptochrome/photolyase, N-terminal domain; |
| sequence family | FAD binding domain of DNA photolyase; |
| redundant complexes |  5zcw_B 5zcw_B
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | HRWRR |
Estimated binding specificities ?
| contact | 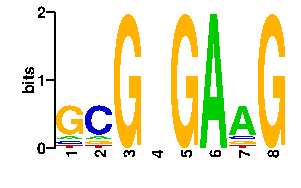 |
A | 9 9 0 24 0 96 67 0 C | 9 67 0 24 0 0 11 0 G | 69 11 96 24 96 0 9 96 T | 9 9 0 24 0 0 9 0scan! |
Dendrogram of similar interfaces ?
matrix format--HGRTNTET--WT--YTMTWTRCWA--RTRG L +-------2xrz_B +--------------------2 ----RT----ETWTYT----------PA---- ! ! +--1tez_B --3 +----1 ----------------------------RG-- L ! +--2wq7_A ! RG------------------------------ +----------------------------4tmu_A |
home
updated Tue Dec 19 07:05:31 2023
