| structure name | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH LEFT END 26-MER DNA (TRANSPOSASE ORFA) | ||||
| reference | Ronning et al. 'Cell(Cambridge,Mass.)' 132 208 2008 | ||||
| source | HELICOBACTER PYLORI | ||||
| experiment | X-ray (resolution=2.10, R-factor=0.225) | ||||
| structural superfamily | Transposase IS200-like; | ||||
| sequence family | Transposase IS200 like; | ||||
| multimeric complexes | 2a6o_AB 2vhg_AB 2vic_AB 2vih_AB 2vju_AB | ||||
| redundant complexes | 2a6o_B
2vhg_A
2vic_A
 2vju_A 2vju_A
| ||||
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| contact | 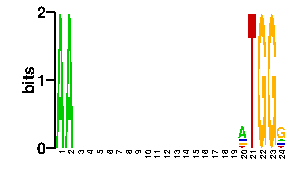 |
A | 96 96 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 54 0 0 0 13 C | 0 0 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 13 0 0 0 13 G | 0 0 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 13 0 96 96 54 T | 0 0 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 16 96 0 0 16scan! |
Dendrogram of similar interfaces ?
matrix formathome
updated Tue Dec 19 06:59:02 2023
