| structure name | CRYSTAL STRUCTURE OF HOXA9 AND PBX1 HOMEODOMAINS BOUND TO DNA (PRE-B-CELL LEUKEMIA TRANSCRIPTION FACTOR-1) |
| reference | Laronde-Leblanc et al. 'Genes 17 2060 2003 |
| source | HOMO SAPIENS |
| experiment | X-ray (resolution=1.90, R-factor=0.233) |
| structural superfamily | Homeodomain-like; |
| sequence family | Homeobox domain; |
| multimeric complexes | 1b72_AB 1b8i_AB 1puf_AB 2r5y_AB 2r5z_AB 4cyc_AB 4uus_AB 4uus_EF 5zjq_AB 5zjr_AB 5zjs_AB |
| redundant complexes |  1lfu_P
5zjr_B
1b72
1b8i
2r5y
2r5z
4cyc
4uus
5zjq
5zjs 1lfu_P
5zjr_B
1b72
1b8i
2r5y
2r5z
4cyc
4uus
5zjq
5zjs
|
| links to other resources | NAKB  PDIdb DNAproDB PDIdb DNAproDB |
| protein sequence | |
| interface signature | RNNIR |
Estimated binding specificities ?
| readout + contact | 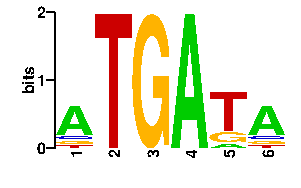 |
A | 72 0 0 96 10 69 C | 8 0 0 0 8 11 G | 8 0 96 0 17 8 T | 8 96 0 0 61 8scan! |
Related DNA sequences reported in the literature ?
| site | source | matches (E-value) |
|---|---|---|
| term: PBX1 | ||
| ATCAATCAA | PubMed | 1puf_B(1.14e-02), 1puf_AB(1.49e-03) |
| TGATTGAC | PubMed | 1puf_B(7.15e-03), 1puf_AB(7.14e-07) |
| TGATTGAT | PubMed | 1puf_B(7.15e-03), 1puf_AB(3.42e-04) |
| TTGAT | PubMed | 1puf_B(4.18e-04), 1puf_AB(7.82e-03) |
| TGATTAAT | PubMed | 1puf_B(7.15e-03), 1puf_AB(3.42e-04) |
| TTGATTGAT | PubMed | 1puf_B(1.14e-02), 1puf_AB(1.49e-03) |
| ATCAATCA | PubMed | 1puf_B(7.15e-03), 1puf_AB(3.42e-04) |
Dendrogram of similar interfaces ?
matrix formatRCRT------------------NT--NAITRG------------ L +----5zjt_B +------------------5 --RA------------------ITQTNAMG-------------- L ! +----5zjt_E ! --RT------------------NT--NAIARG------------ L ! +----1b72_B ! +------4 ----------------------NCITNA----------RGRG-- L ! ! +----6fqq_E ! +--7 --RT----------------KTIAQCNA----RT---------- L ! ! ! +1ig7_A ! ! +----------2 --RT------------------ITQCNAACRG------------ L ! ! +6a8r_A +-12 +-8 --RT------------------NT--NA--RG------------ L ! ! ! ! +-1b8i_B ! ! ! ! +--------3 --RT------------------IAKGNA---------------- L ! ! ! ! ! ! +2hdd_A ! ! +-9 +---6 +-1 ----------------------IA--NA---------------- ! ! ! ! ! +4qtr_D ! ! ! ! ! --------------YTVC--KGDCKC------------------ L -13 ! +-10 ! +----------2qhb_A ! ! ! ! ! --RA------YA----------IAQCNAYCKG------------ L ! ! ! ! +----------------5flv_I ! +--11 ! ----------------RTQT--ITQANA------VTKT----KT L ! ! +-----------------2ld5_A ! ! ----NGRGTG--LG--LCRG------------------------ ! +-------------------4e9f_A ! --RA----------------KGSAQA------------------ L +-------------------------6j5b_C |
home
updated Tue Dec 19 06:02:14 2023
