matrix-quality result: PurR_2pua_A
Command:
matrix-quality -v 1 -m /export/internal_use2/amedina/matrix_eval/data/Matrices_contact/PurR/PurR_2pua_A.tab -matrix_format tab -seq db_sites /export/internal_use2/amedina/matrix_eval/data/Sites_FNA_R/PurR.fna -perm db_sites 0 -scanopt db_sites '-uth rank_pm 1' -seq allup-noorf /export/internal_use2/amedina/matrix_eval/data/sequences/allup/Escherichia_coli_K12_allup-noorf_min30_max500.fasta -perm allup-noorf 1 -roc_ref theor -img_format png,pdf -graph_option '-title1 PurR' -graph_option '-title2 /export/internal_use2/amedina/matrix_eval/data/Matrices_contact/PurR/PurR_2pua_A.tab' -graph_option '-xgstep1 5 -xgstep2 1' -graph_option '-ymin 1.0e-7 -ymax 1' -graph_option '-xmin -50 -xmax 30' -roc_option '-gstep1 0.1 -gstep2 0.05' -roc_option '-title1 PurR' -roc_option '-title2 /export/internal_use2/amedina/matrix_eval/data/Matrices_contact/PurR/PurR_2pua_A.tab' -2str -pseudo 1 -bgfile /export/space2/rsa-tools/data/genomes/Escherichia_coli_K12/oligo-frequencies/1nt_upstream-noorf_Escherichia_coli_K12-ovlp-1str.freq -th_prior /export/space2/rsa-tools/data/genomes/Escherichia_coli_K12/oligo-frequencies/1nt_upstream-noorf_Escherichia_coli_K12-ovlp-1str.freq -o /export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A.tab_quality -graph_option '-colors /export/internal_use2/amedina/matrix_eval/data/quality_colors.tab ' -roc_option '-colors /export/internal_use2/amedina/matrix_eval/data/quality_colors.tab'
Figures
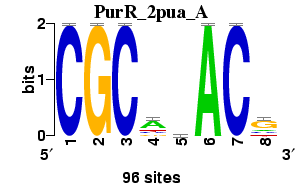
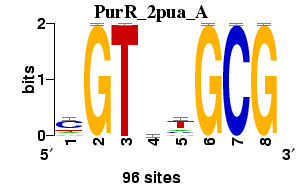
Matrix logo
Complementary cumulative distributions
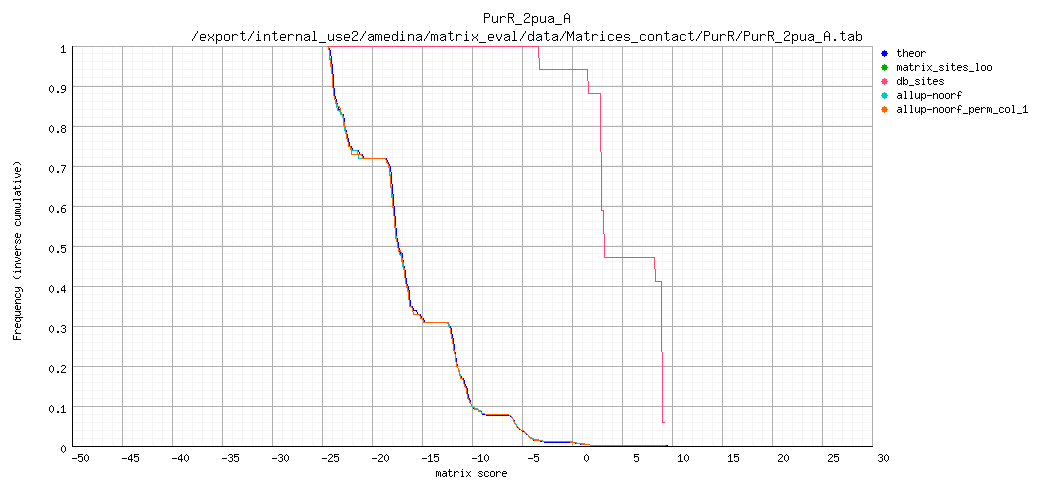
Complementary cumulative distributions (logarithmic Y axis)
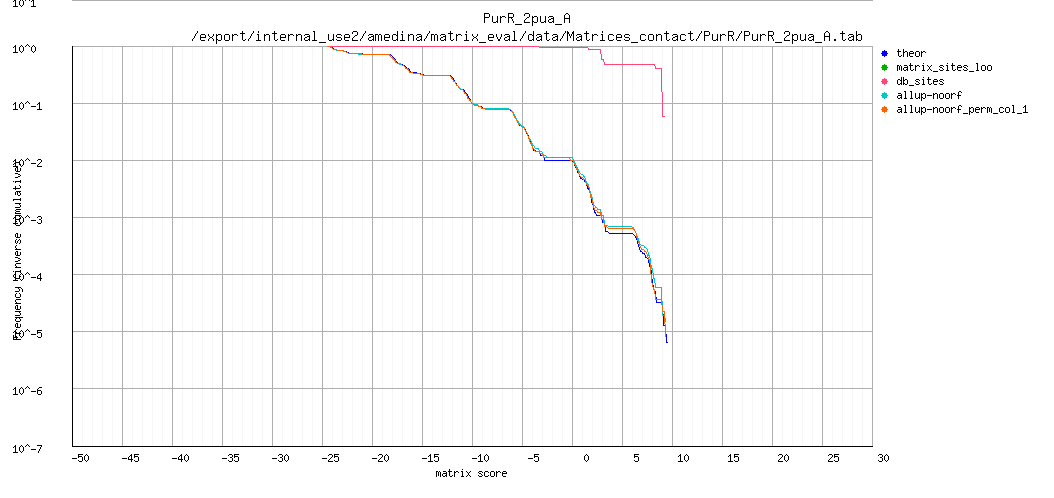
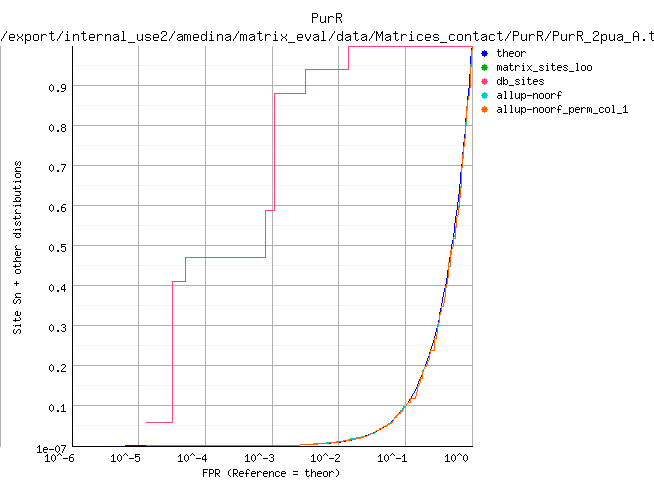
ROC curve (logarithmic X axis)
Matrix information
; convert-matrix -v 1 -i /export/internal_use2/amedina/matrix_eval/data/Matrices_contact/PurR/PurR_2pua_A.tab -from tab -to tab -o /export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A.tab_quality_matrix_info.txt -bgfile /export/space2/rsa-tools/data/genomes/Escherichia_coli_K12/oligo-frequencies/1nt_upstream-noorf_Escherichia_coli_K12-ovlp-1str.freq -bg_format oligos -return counts,frequencies,weights,info,parameters,sites,logo -logo_format png,pdf -logo_opt '-e -M -t PurR_2pua_A '
; Input files
; input /export/internal_use2/amedina/matrix_eval/data/Matrices_contact/PurR/PurR_2pua_A.tab
; prior /export/space2/rsa-tools/data/genomes/Escherichia_coli_K12/oligo-frequencies/1nt_upstream-noorf_Escherichia_coli_K12-ovlp-1str.freq
; Input format tab
; Output files
; output /export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A.tab_quality_matrix_info.txt
; Output format tab
; pseudo-weight 1
; Background model
; Bernoulli model (order=0)
; Strand sensitive
; Background pseudo-frequency 0.01
; Residue probabilities
; a 0.29107
; c 0.20737
; g 0.20436
; t 0.29721
a | 0 0 0 54 24 96 0 16
c | 96 0 96 13 24 0 96 13
g | 0 96 0 13 24 0 0 54
t | 0 0 0 16 24 0 0 13
//
a | 0.0 0.0 0.0 0.6 0.3 1.0 0.0 0.2
c | 1.0 0.0 1.0 0.1 0.2 0.0 1.0 0.1
g | 0.0 1.0 0.0 0.1 0.2 0.0 0.0 0.6
t | 0.0 0.0 0.0 0.2 0.3 0.0 0.0 0.1
//
a | -4.6 -4.6 -4.6 0.7 -0.2 1.2 -4.6 -0.5
c | 1.6 -4.6 1.6 -0.4 0.2 -4.6 1.6 -0.4
g | -4.6 1.6 -4.6 -0.4 0.2 -4.6 -4.6 1.0
t | -4.6 -4.6 -4.6 -0.6 -0.2 -4.6 -4.6 -0.8
//
a | -0.0 -0.0 -0.0 0.4 -0.0 1.2 -0.0 -0.1
c | 1.6 -0.0 1.6 -0.1 0.0 -0.0 1.6 -0.1
g | -0.0 1.6 -0.0 -0.1 0.0 -0.0 -0.0 0.6
t | -0.0 -0.0 -0.0 -0.1 -0.0 -0.0 -0.0 -0.1
//
; Sites
;
; Matrix parameters
; Columns 8
; Rows 4
; Alphabet a|c|g|t
; Prior a:0.291066898814656|c:0.207372933914605|g:0.204355073413807|t:0.297205093856932
; program tab
; matrix.nb 1
; pseudo 1
; info.log.base 2.71828
; min.prior 0.204355
; alphabet.size 4
; max.bits 2
; total.information 7.73774
; information.per.column 0.967217
; max.possible.info.per.col 1.5879
; consensus.strict CGCagACG
; consensus.strict.rc CGTCTGCG
; consensus.IUPAC CGCasACG
; consensus.IUPAC.rc CGTSTGCG
; consensus.regexp CGCa[cg]ACG
; consensus.regexp.rc CGT[CG]TGCG
; residues.content.crude.freq a:0.2474|c:0.4401|g:0.2435|t:0.0690
; G+C.content.crude.freq 0.683594
; residues.content.corrected.freq a:0.2478|c:0.4377|g:0.2431|t:0.0714
; G+C.content.corrected.freq 0.680791
; min(P(S|M)) 1.94781e-16
; max(P(S|M)) 0.0752577
; proba_range 0.0752577
; Wmin -24.5
; Wmax 9.5
; Wrange 34
; logo file:/export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A_m1_logo.png
; logo file:/export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A_m1_logo.pdf
; logo file:/export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A_m1_logo_rc.png
; logo file:/export/internal_use2/amedina/matrix_eval/results/contact/matrix_quality_200100311/PurR/PurR_2pua_A.tab/PurR_2pua_A_m1_logo_rc.pdf
; Job started 2010_03_11.201143
; Job done 2010_03_11.201147
Result files




